In the rapidly evolving field of protein engineering, a groundbreaking approach called EVOLVEpro is making waves. This innovative system combines the power of protein language models (PLMs) with active learning techniques to achieve remarkable improvements in protein properties while minimizing laboratory experimentation. Let’s delve into how EVOLVEpro is transforming the landscape of protein engineering and its potential impact on various applications.
Protein engineering aims to modify proteins to enhance their properties or create entirely new functions. However, traditional methods face significant challenges: the vast sequence space makes it difficult to identify beneficial mutations, conventional directed evolution techniques can be time-consuming and resource-intensive, and zero-shot predictions from PLMs often yield limited success in real-world applications.
EVOLVEpro addresses these challenges by integrating two powerful concepts: Protein Language Models (PLMs) and Active Learning. PLMs are AI models that capture broad patterns in protein structure and function based on evolutionary data. Active Learning is an approach that strategically selects a small set of data points for experimental testing to maximize information gain.
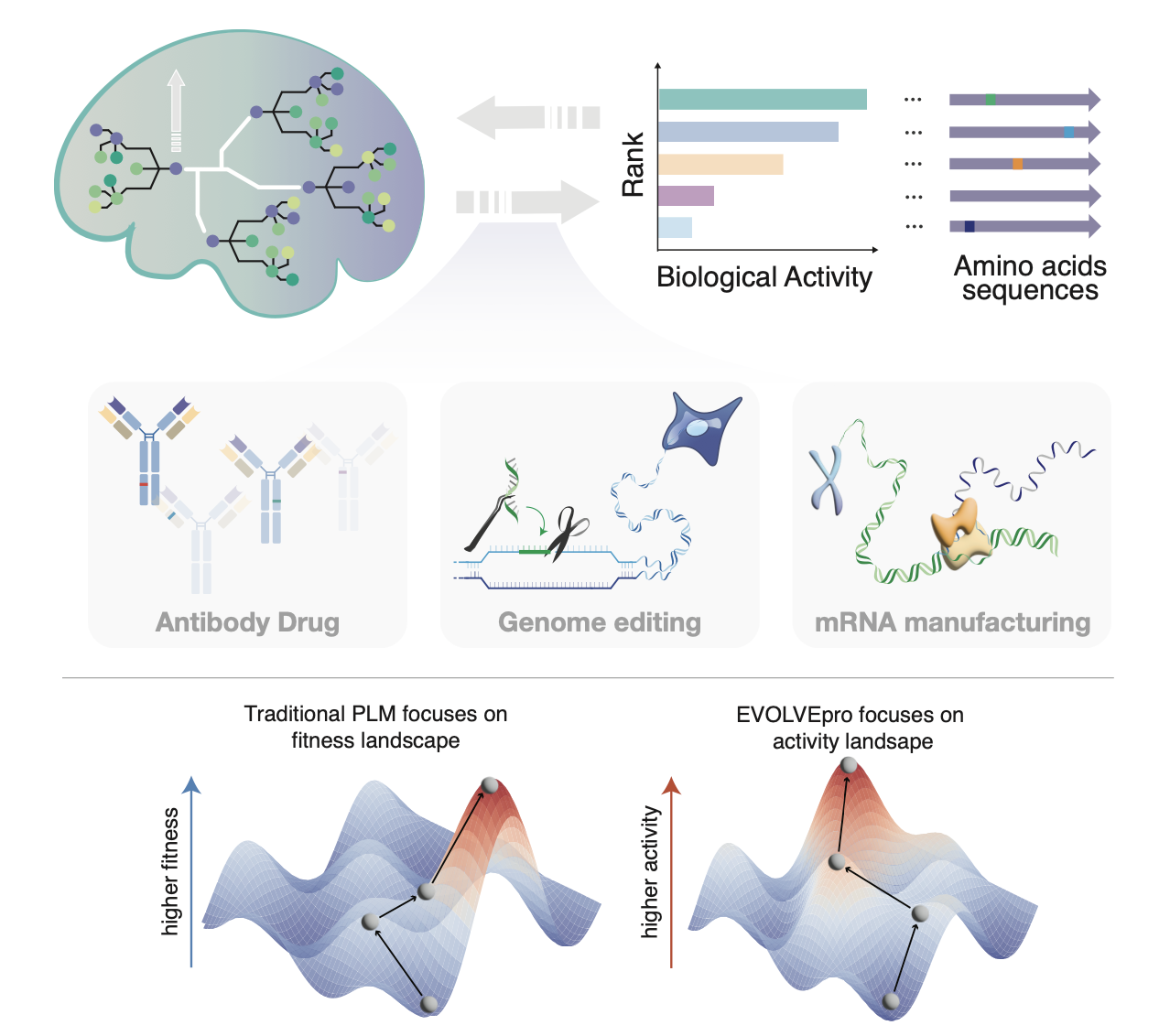
EVOLVEpro’s few-shot learning framework requires minimal experimental data to optimize diverse proteins. It efficiently navigates fitness landscapes, avoiding local optima that often trap traditional methods. The system can enhance multiple protein properties simultaneously and has been successfully tested on various proteins, including CRISPR nucleases, antibodies, and RNA polymerases.
EVOLVEpro has demonstrated its effectiveness in several critical areas of protein engineering. In CRISPR nuclease engineering, it evolved PsaCas12f, a compact CRISPR nuclease, to achieve 50% editing efficiency at mammalian genomic loci, outperforming state-of-the-art miniature nucleases and opening new possibilities for gene editing applications. In antibody engineering, EVOLVEpro showcased multi-objective optimization by enhancing both binding affinity and expression levels of antibodies, demonstrating its potential for accelerating therapeutic antibody design and development. In RNA polymerase optimization, the system engineered T7 RNA polymerase (RNAP) mutants with increased RNA production fidelity and reduced immunogenic byproducts, advancing applications in RNA therapeutics.
EVOLVEpro’s success lies in its unique architecture, which integrates a PLM with a regression layer to combine broad protein knowledge with the ability to learn specific activity landscapes. The system uses active learning to iteratively select small sets of mutations for testing, continuously improving predictions. Notably, EVOLVEpro operates solely on protein sequences, eliminating the need for structural information or expert knowledge.
The developers of EVOLVEpro rigorously tested the system across 12 diverse datasets to ensure broad applicability. The system achieved up to 515-fold improvements in desired properties compared to conventional methods and demonstrated the ability to select highly active mutants from vast sequence spaces, such as single mutants from over 16,000 possibilities and multi-mutants from over 780 billion options.
EVOLVEpro’s success opens up exciting possibilities for the future of protein engineering. It promises accelerated drug discovery through faster optimization of therapeutic proteins and antibodies, enhanced biotechnology tools with improved enzymes and molecular biology reagents, sustainable solutions through engineered proteins for environmental and industrial applications, and personalized medicine with tailored protein therapeutics based on individual genetic profiles.
In conclusion, EVOLVEpro represents a significant leap forward in protein engineering, bridging the gap between computational predictions and experimental validation. By combining the power of AI with strategic experimental design, it offers a more efficient and effective approach to protein optimization. As this technology continues to evolve and integrate with next-generation protein language models, we can expect even more remarkable advancements in the field, potentially revolutionizing areas from medicine to biotechnology and beyond.
Paper | Science







Post comments