
Research identifies key cellular mechanisms driving Alzheimer's progression
Scientists at Washington University School of Medicine in St. Louis have established connections between disease-associated proteins and genes to uncover specific cellular pathways involved in Alzheimer's development and advancement. The proteins, extracted from cerebrospinal fluid, effectively represent brain activity. Several of these proteins could serve as potential therapeutic targets.
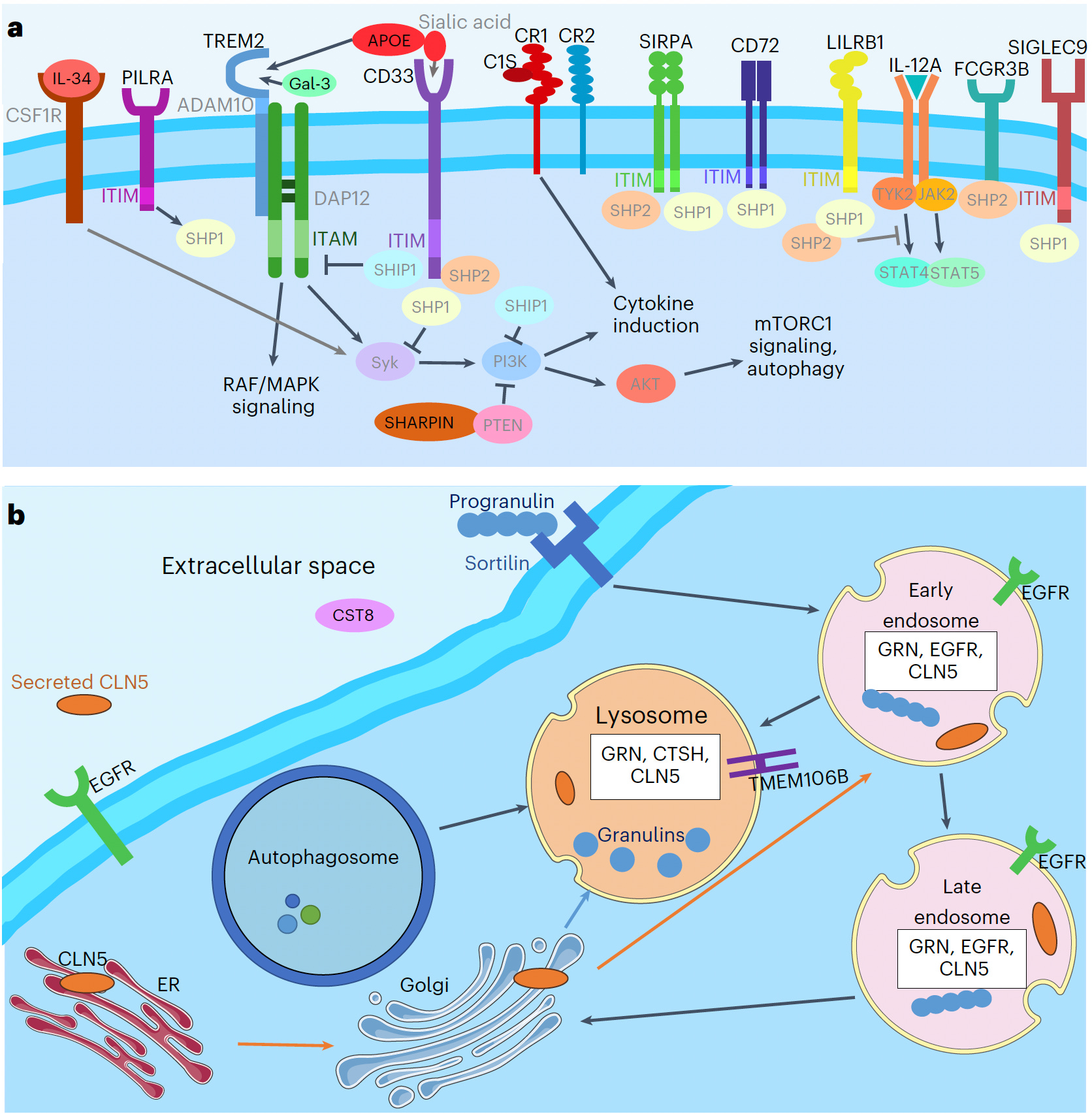
Scientists have made a breakthrough in understanding Alzheimer's disease by analyzing cerebrospinal fluid (CSF) from living patients. For the first time, researchers at Washington University School of Medicine in St. Louis have successfully mapped the relationship between disease-related proteins and genes, identifying specific cellular pathways crucial to the onset and progression of Alzheimer's. These proteins, collected from CSF, serve as reliable indicators of brain activity, and several show promise as potential therapeutic targets. The research findings have been published in Nature Genetics.
According to Carlos Cruchaga, PhD, the Barbara Burton and Reuben Morriss III professor of psychiatry and director of the NeuroGenomics and Informatics Center at WashU Medicine, utilizing patients' CSF represents a significant advancement in such research and may be the optimal method for obtaining relevant samples to map protein activity patterns, known as the proteome.
"Our primary objective is to identify both risk-associated and protective genes, while also understanding their causal roles," stated Cruchaga. "This requires studying human-derived data, which led us to conduct an extensive proteomic analysis of cerebrospinal fluid, as CSF effectively reflects the disease's pathology."
Cruchaga noted that previous studies relied on post-mortem brain tissue, limiting insights to late-stage Alzheimer's. Other research examined blood plasma, which lacks specificity to disease-affected tissues.
Over the past fifteen years of Alzheimer's research, scientists have expanded the number of genome regions linked to the condition from 10 to approximately 80. However, identifying associated genes or DNA regions is just the beginning. Connecting an individual's proteomic profile – the active proteins and their levels – to their genetic code provides a comprehensive view of brain cellular activities.
By analyzing CSF samples from individuals with and without Alzheimer's, researchers could identify malfunctioning cellular pathways.
"Within DNA regions associated with Alzheimer's, we often find multiple genes, and it's unclear which ones drive the condition," Cruchaga explained. "Including proteins in our analysis helps identify the driving genes, determine their molecular pathways, and reveal previously undiscovered protein interactions."
Through the Knight-ADRC and the Dominantly Inherited Alzheimer Network (DIAN) at WashU Medicine, along with collaborative studies, Cruchaga's team accessed an extensive database. These studies provided genetic information and CSF samples from 3,506 participants, including both healthy donors and Alzheimer's patients.
The researchers compared proteomic data from CSF samples with existing studies identifying Alzheimer's-related genome areas. This analysis focused on 1,883 proteins from the 6,361 in the CSF proteomic atlas. Using three established statistical methods, they confidently identified genes and proteins within disease-related biological pathways.
This approach revealed 38 proteins likely causing Alzheimer's progression, with 15 being potential drug targets.
"Our analysis uniquely identifies risk-modifying proteins," said Cruchaga. "Understanding these causal steps allows us to trace their effects in the brain."
While this study's immediate impact on understanding and treating Alzheimer's is substantial, Cruchaga believes CSF proteomics could reveal crucial information about various neurological conditions, from Parkinson's disease to schizophrenia.
"This approach's strength lies in its versatility – once you have mapped genetic variants and protein levels, you can apply it to any disease," he explained.
Beyond proteins, Cruchaga is exploring metabolites in CSF – substances released during cellular processes. In a separate Nature Genetics publication, he and his colleagues demonstrated this approach's potential, reporting connections between specific metabolites and conditions including Parkinson's disease, diabetes, and dementia.
Source: https://genomemedicine.biomedcentral.com/articles/10.1186/s13073-024-01388-3




Post comments