The intricate relationship between the gut microbiome and rheumatoid arthritis (RA) has become a significant area of research, with recent studies providing valuable insights into how gut bacteria might influence the onset and progression of this chronic autoimmune disease. RA, characterized by joint inflammation and potential long-term damage, is thought to arise from a combination of genetic predisposition and environmental factors. In recent years, the role of the gut microbiome—comprising billions of microorganisms in the digestive tract—has garnered attention as a potential contributor to RA development. This article summarizes the findings of a groundbreaking study that explores gut microbiome dynamics in individuals at risk of RA, focusing on how microbial changes might serve as early indicators of the disease.
Background on Rheumatoid Arthritis and Gut Microbiome
RA is an autoimmune disorder where the immune system mistakenly attacks the joints, leading to inflammation, pain, and stiffness. While the exact cause remains unknown, it is believed that genetic factors combined with environmental triggers, including infections or diet, play a role in disease initiation. The gut microbiome, which plays a critical role in immune regulation, is thought to contribute to autoimmune diseases like RA. A healthy gut microbiome helps modulate immune responses and reduces systemic inflammation, while dysbiosis (microbial imbalance) can lead to the development of inflammatory diseases, including RA.
Study Design and Methodology
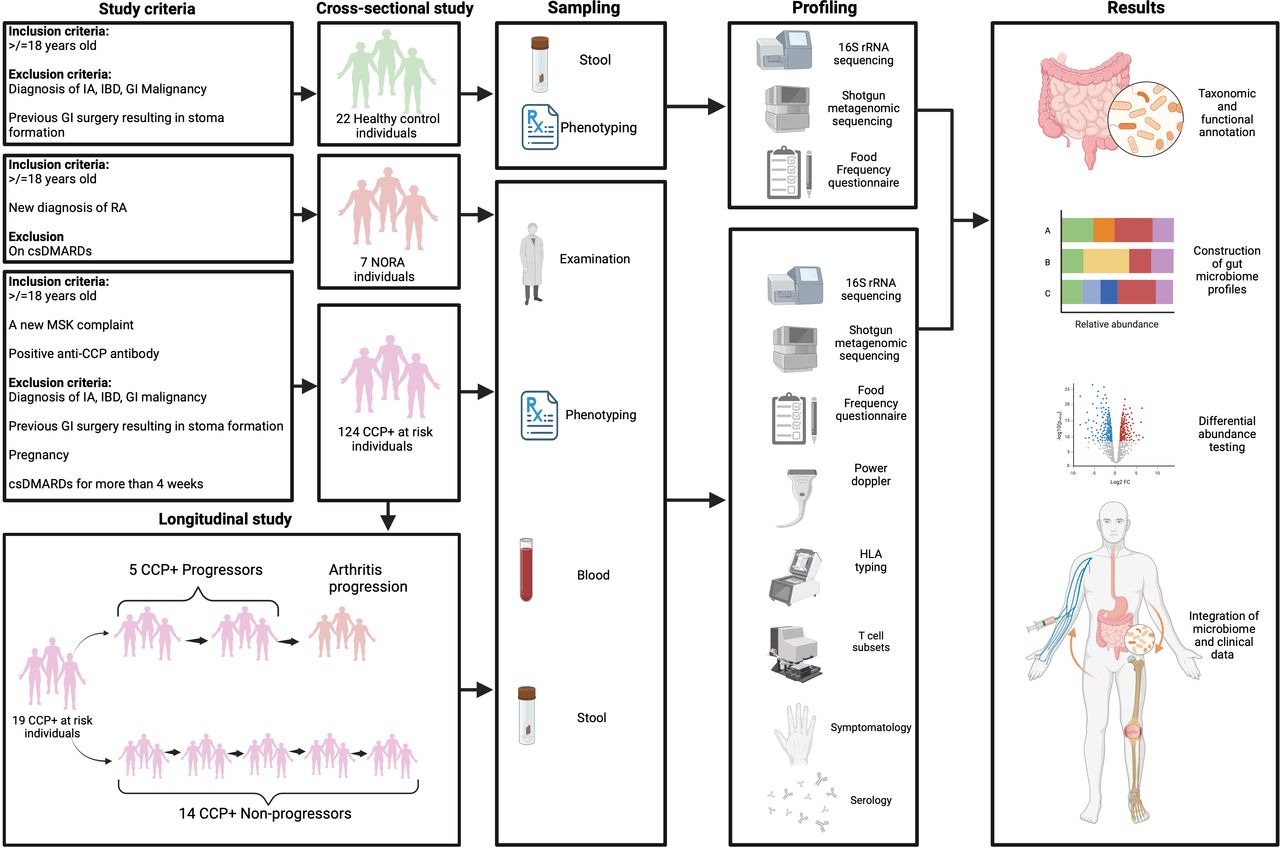
The study employed a multi-faceted approach combining cross-sectional and longitudinal analyses to capture a comprehensive picture of gut microbiome dynamics in RA. Participants included individuals at high risk for RA, those with newly diagnosed RA (NORA), and healthy controls. The high-risk individuals were identified through the presence of anti-cyclic citrullinated peptide (anti-CCP) antibodies and early musculoskeletal symptoms, while NORA participants had just received an RA diagnosis. Stool samples were collected from all participants for microbiome analysis, and advanced sequencing techniques were used to identify microbial composition and functional changes:
16S rRNA sequencing provided broad taxonomic classifications of the gut bacteria.
Shotgun metagenomic sequencing allowed for more detailed strain-level analysis and functional profiling.
This methodology enabled researchers to identify specific microbial changes and how they correlated with RA development.
Key Findings: Changes in Gut Microbiome Composition
The study's findings revealed significant differences in the gut microbiomes of individuals at high risk for RA compared to healthy controls. Most notably, participants at high risk showed reduced gut microbiome diversity. A more diverse microbiome is generally associated with better health outcomes, so this loss of diversity is concerning. The study also identified specific changes in bacterial abundance, particularly within the Prevotellaceae family. These changes varied in a strain-specific manner, with some strains more abundant in high-risk individuals and others less so. This suggests that not all Prevotellaceae strains play the same role in RA risk, which complicates previous assumptions about the microbiome's involvement in the disease.
Temporal Dynamics: Microbiome Changes Leading to RA Onset
The longitudinal aspect of the study was particularly enlightening. By analyzing microbiome samples over time, researchers observed that microbial changes could precede the clinical onset of RA. These shifts in the microbiome appeared to serve as early indicators of disease, suggesting that gut dysbiosis might occur before visible symptoms of RA appear. This finding is significant as it presents the possibility of using microbiome analysis as a diagnostic tool to identify individuals at risk for RA before clinical symptoms manifest. Early intervention strategies, such as dietary or lifestyle modifications to promote a healthier microbiome, could potentially delay or prevent the onset of RA.
Functional Changes in the Gut Microbiome
In addition to structural changes in the microbiome, the study also examined functional alterations associated with RA risk. Using shotgun metagenomics, the researchers identified specific metabolic pathways that were more active in individuals with newly diagnosed RA. Notably, there was an increase in pathways related to amino acid metabolism, particularly those involving lysine and threonine, as well as pathways involved in energy production and the tricarboxylic acid (TCA) cycle. These functional changes likely reflect metabolic shifts associated with inflammation, providing further evidence that the gut microbiome influences immune responses and inflammation in RA.
Implications for RA Pathogenesis and Future Research
The study offers several important insights into RA pathogenesis:
Dysbiosis as an Early Event: The observed decrease in microbiome diversity in high-risk individuals suggests that dysbiosis may be an early event in RA development, potentially occurring before the onset of clinical symptoms.
Strain-Specific Changes: The identification of specific bacterial strains within families like Prevotellaceae that are associated with RA risk highlights the need for more detailed, strain-level analysis in microbiome studies. Broad taxonomic classifications may overlook important nuances that could be crucial for understanding disease mechanisms.
Metabolic Pathway Alterations: The changes in metabolic pathways identified in RA patients point to possible mechanisms by which the gut microbiome could influence immune function and inflammation. These pathways could provide new targets for therapeutic intervention.
These findings open up several avenues for future research, including the investigation of specific mechanisms by which gut bacteria contribute to RA, the development of microbiome-based biomarkers for early detection, and the potential for microbiome-modulating therapies to prevent or treat RA.
Limitations and Future Directions
While the study provides valuable insights, it also has limitations. The relatively small sample size, particularly for the longitudinal analysis, means that the findings may not be universally applicable. The study also focused on bacterial components of the microbiome, leaving the roles of viruses, fungi, and other microorganisms unexplored. Additionally, the lack of integrated data on host gene expression and metabolomics limits the ability to draw definitive conclusions about the functional consequences of microbiome changes.
Future research should aim to address these limitations by conducting larger, more diverse studies and incorporating multi-omics approaches. Integrating genomic, transcriptomic, and metabolomic data will provide a more comprehensive understanding of how the microbiome influences RA development. Mechanistic studies using animal models and human cell cultures will be essential to establish causal links between specific microbial changes and RA.
Conclusion
This study represents a significant step forward in understanding the relationship between the gut microbiome and rheumatoid arthritis. By using advanced sequencing techniques and a multi-phase study design, the researchers have provided new insights into the microbial changes that precede and accompany RA development. The identification of specific bacterial strains and metabolic pathways linked to RA risk opens the door for new diagnostic tools and therapeutic strategies aimed at modulating the microbiome. As research in this area continues to evolve, microbiome-based interventions could play a key role in preventing and managing rheumatoid arthritis, offering a promising avenue for future treatment.
Source: https://ard.bmj.com/content/early/2024/10/22/ard-2024-226362






Post comments