Deep generative modeling has received much attention in the field of ab initio drug design. However, rational design of ligand molecules for novel targets remains challenging, especially in controlling the properties of the generated molecules.
Here, inspired by DNA-encoded compound library technology, researchers propose DeepBlock, a block (block)-based deep learning approach for ligand generation that can be tailored to target protein sequences while enabling precise property control.
In addition, DeepBlock can combine optimization algorithms and deep learning to regulate the properties of the generated molecules.
The study, titled “A deep learning approach for rational ligand generation with toxicity control via reactive building blocks,” was published on November 8, 2024, in Nature Computational Science.

Finding small-molecule ligands that bind to specific proteins is a critical part of drug discovery. Virtual screening has emerged as an important method for identifying biologically active compounds in small molecule libraries using computer programs. However, the effectiveness of virtual screening is limited by the vast chemical space and the library of compounds used.
In contrast, ab initio drug design strategies (generating molecular structures from scratch) offer a promising avenue for exploring a wider chemical space beyond existing libraries.
In recent years, these models have made significant progress in molecule generation, but they typically lack the ability to generate for specific protein targets, and thus need to be compensated for by additional screening or by incorporating techniques such as reinforcement learning. In addition, the synthesizability of the generated molecules and the toxicity and metabolism of the drugs need to be considered in the actual drug development.
DNA-encoded compound library technology has become a widely accepted wet-lab drug discovery method. This method utilizes combinatorial chemistry to rapidly generate a large number of candidate compounds through the reaction of molecular building blocks.
DeepBlock
Inspired by the DNA-encoded compound library technique, a team of researchers from Hunan University and Xi'an University of Electronic Science and Technology proposed DeepBlock, a deep learning-based framework that utilizes molecular building blocks for ab initio drug design. Building blocks in this context represent molecular fragments that are capable of reacting with each other chemically.
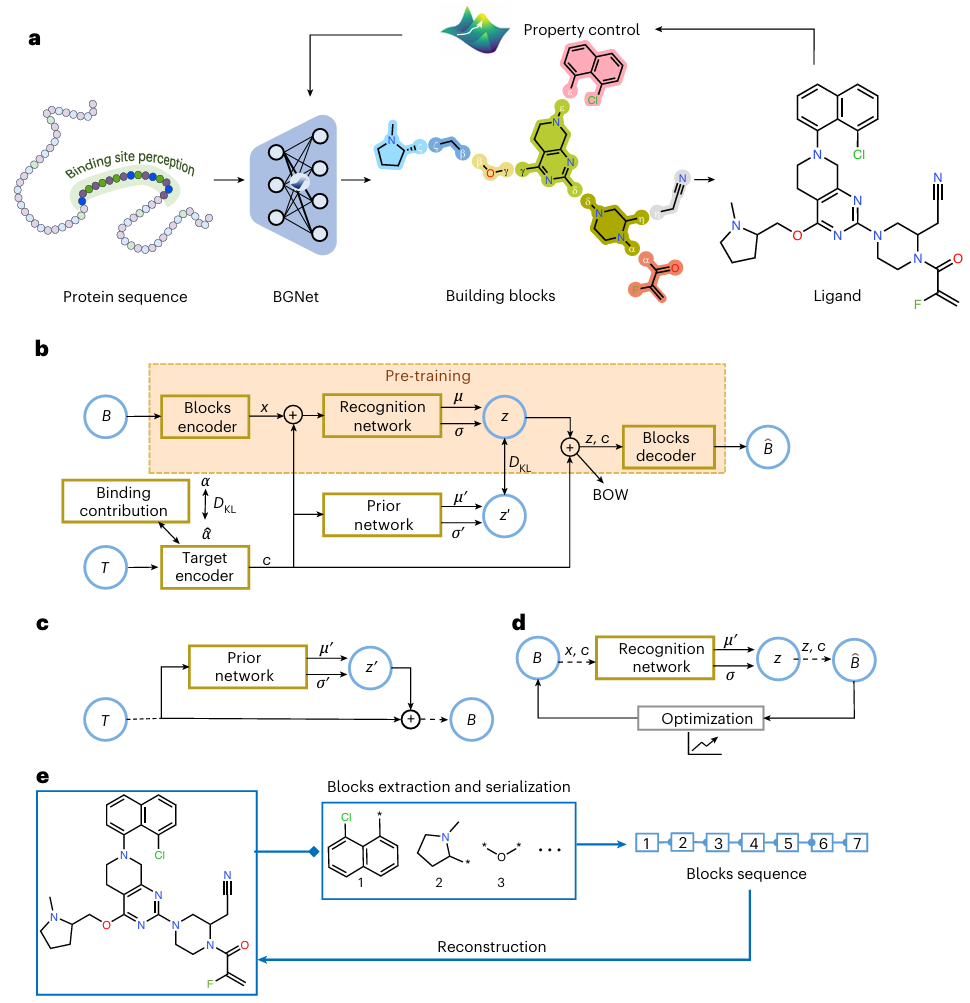
Icon: Overview of the DeepBlock framework.
The core idea of DeepBlock is to decompose the molecule generation process into two consecutive steps: first, generating building blocks based on protein sequence embedding features, and then assembling them into complete molecules. By exploiting the inherent properties of these blocks and the chemical interactions between them, DeepBlock can design better quality rational molecules.
Based on this concept, the researchers designed effective mechanisms in DeepBlock to address two key tasks: customizing molecule generation to protein sequences and property control during generation.
In DeepBlock, the team incorporated the Block Generation Network (BGNet), a conditional deep generation model designed to generate block sequences based on a given protein sequence.BGNet combines two key features that significantly improve its performance.
First, it is built from a molecular block autoencoder pre-trained on a large-scale molecular dataset containing an extensive lexicon of 10,701 blocks containing a variety of commonly used fragments. This pre-training extends the chemical space and mitigates potential overfitting due to the limited size of the protein-ligand pair dataset.
Second, the researchers introduced a key component in DeepBlock, the Target Contribution Awareness Module. This module enhances the model's ability to recognize ligand-residue interactions autonomously and compensates for the lack of 3D structural information in protein sequences. The combination of these two features in BGNet highlights its ability to generate diverse and biologically active molecular fragments, effectively addressing the challenges posed by protein sequence data.
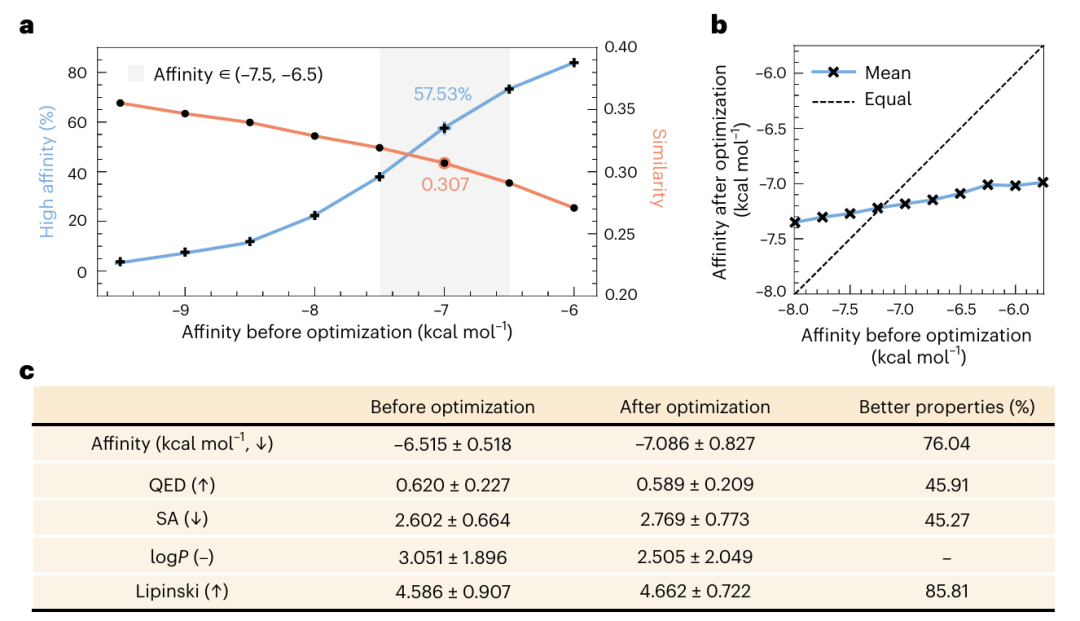
Illustration: affinity comparison before and after optimization.
In addition, the team used BGNet in conjunction with the Simulated Annealing (SA) algorithm or Bayesian Optimization (BO) to control the generation process, aiming to enhance the other features while retaining its binding affinity for the target protein.

Illustration: optimization process and results.
The research team also conducted experiments with drug toxicity as the optimization goal. When combined with simulated annealing or Bayesian optimization optimized for toxicity, DeepBlock successfully generated ligands with low toxicity while retaining affinity for the target.
Future Work
The method also has limitations.
DeepBlock can currently only generate blocks from an existing block dictionary, thus limiting the diversity of molecules it generates. The team's future research direction is to explore methods for generating blocks from scratch, thus freeing the model from the limitations of existing dictionary blocks and unlocking the potential for greater versatility and novelty in the molecules it can create.
In addition, DeepBlock generates two-dimensional (2D) molecular structures as SMILES strings, providing controllable properties and applicability to new targets. While SMILES strings provide sufficient structural information for various drug development scenarios, they lack 3D structural details.
Future research will focus on combining this approach with methods such as LiGAN to develop controlled 3D molecular generation methods based on molecular building blocks. This hybrid approach could combine the strengths of both 2D and 3D drug design methods, potentially improving the efficiency and effectiveness of drug discovery.
Link to paper: A deep learning approach for rational ligand generation with toxicity control via reactive building blocks







Post comments