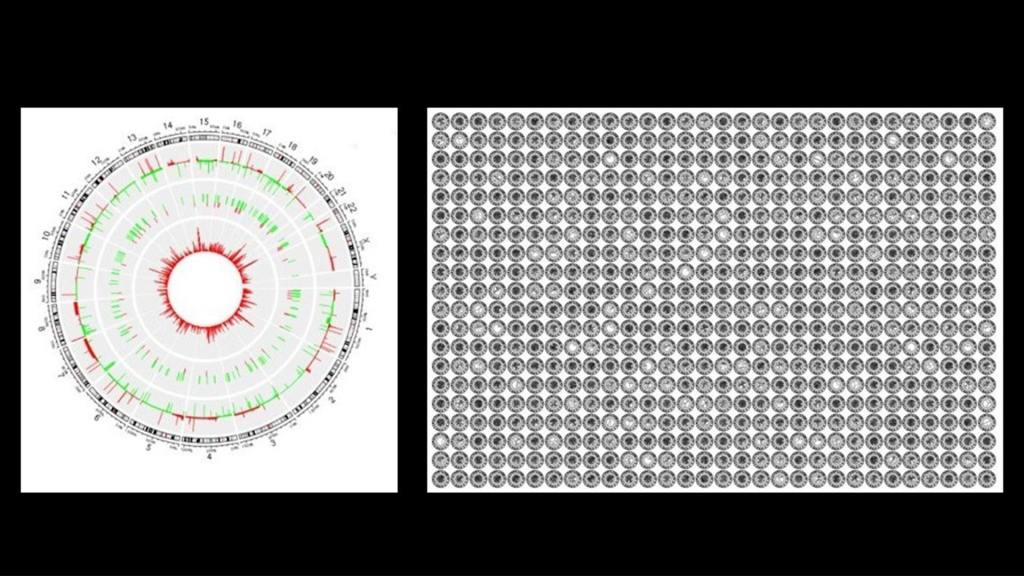
Left: A circular OmicsFootPrint visualization shows genetic and molecular changes, with chromosomes in the outer ring and gene activity changes inside — red for increased activity, green for reduced. Right: A grid of OmicsFootPrints summarizes multi-omics data for nearly 700 cancer patients, with each circle representing a unique molecular profile.
Researchers at Mayo Clinic have developed an artificial intelligence (AI) tool, OmicsFootPrint, designed to convert vast and complex biological data into two-dimensional circular images.
Omics is the study of genes, proteins, and other molecular data to understand how the body functions and how diseases develop. By mapping this data, OmicsFootPrint may provide clinicians and researchers with a novel approach to visualizing disease patterns, such as those found in cancer and neurological disorders, which could help guide personalized therapies. Additionally, the tool offers an intuitive way to explore disease mechanisms and interactions.
The details of this tool have been published in a recent study in Nucleic Acids Research.
Simplifying Complex Data
Genes serve as the body's instruction manual, while proteins execute these instructions to maintain cellular functions. Mutations in these genetic instructions can disrupt these processes, leading to disease. OmicsFootPrint helps interpret these complexities by transforming data—such as gene activity, mutations, and protein levels—into colorful circular maps, providing a clearer picture of biological processes.
In the study, researchers applied OmicsFootPrint to analyze drug response and cancer multi-omics data. The tool demonstrated an 87% average accuracy in distinguishing between two types of breast cancer—lobular and ductal carcinomas. In lung cancer analysis, it exhibited over 95% accuracy in identifying two types: adenocarcinoma and squamous cell carcinoma.
Small Datasets, Big Impact
The study found that integrating multiple types of molecular data yields more accurate results compared to using a single data type.
Furthermore, OmicsFootPrint has demonstrated the ability to generate meaningful results even when working with limited datasets. The tool employs a technique known as transfer learning, an advanced AI method that enables it to learn from existing data and apply that knowledge to new scenarios. In one instance, it achieved over 95% accuracy in identifying lung cancer subtypes while using less than 20% of the typical data volume.
To enhance accuracy and insight, the OmicsFootPrint framework incorporates SHAP (SHapley Additive exPlanations), a method that highlights the most influential biomarkers, genes, or proteins driving disease patterns, helping researchers better understand these factors.
From Research to Clinical Application
Beyond research, OmicsFootPrint has been designed for clinical use. The tool compresses large biological datasets into compact images that require just 2% of the original storage space, facilitating integration into electronic medical records to support future patient care.
The research team aims to expand OmicsFootPrint to study additional diseases, including neurological disorders and other complex conditions. They are also working on updates to enhance the tool’s accuracy and flexibility, including capabilities for identifying new disease markers and drug targets.
For further details, including a full list of authors, disclosures, and funding sources, the complete study is available in Nucleic Acids Research.
Source: Mayo Clinic









Post comments